ozone <- read.table("../donnees/ozone_simple.txt",header=TRUE,sep=";")
plot(O3~T12, data=ozone, xlab="T12", ylab="O3")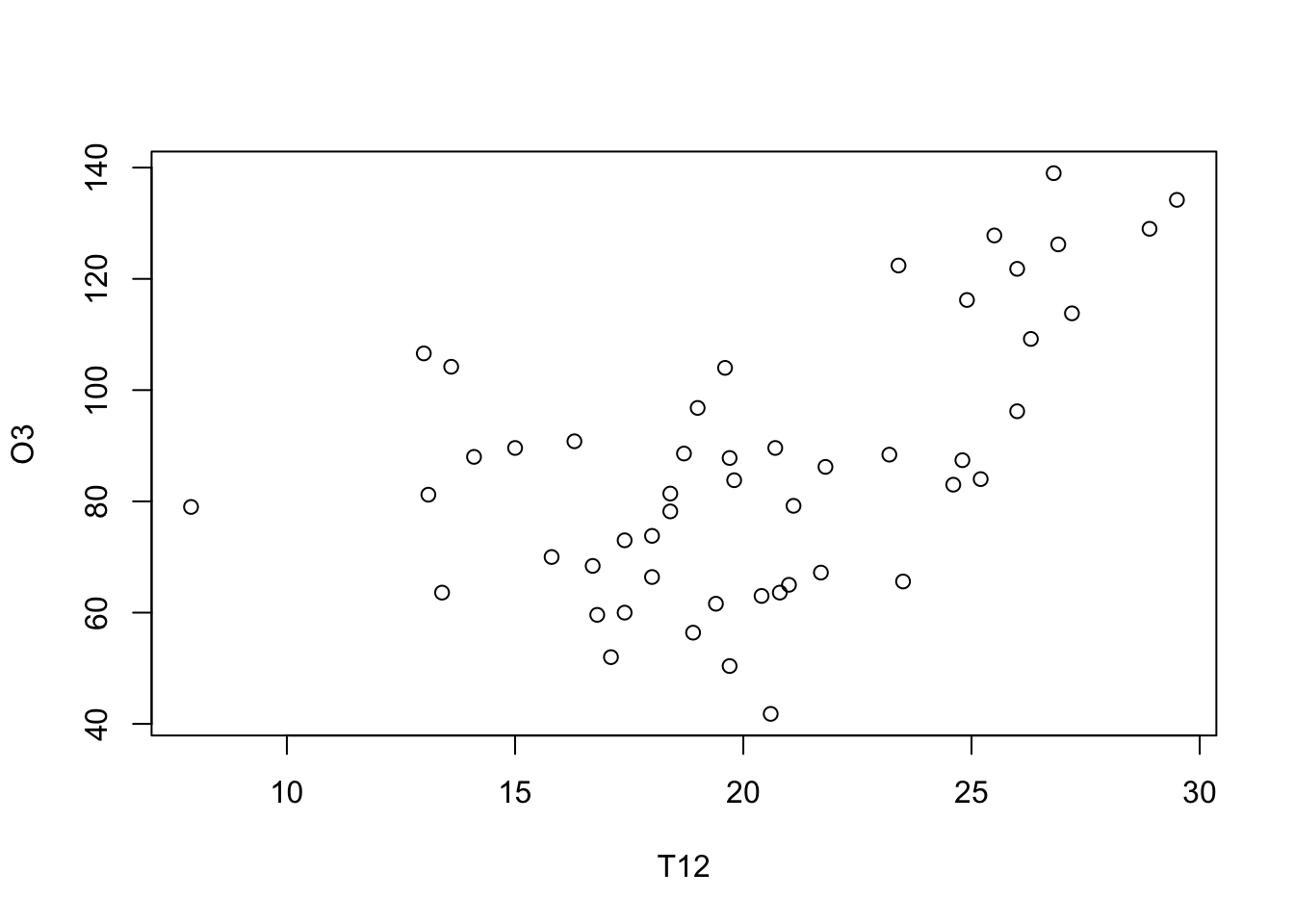
ozone <- read.table("../donnees/ozone_simple.txt",header=TRUE,sep=";")
plot(O3~T12, data=ozone, xlab="T12", ylab="O3")
reg <- lm(O3~T12, data=ozone)
summary(reg)
Call:
lm(formula = O3 ~ T12, data = ozone)
Residuals:
Min 1Q Median 3Q Max
-45.256 -15.326 -3.461 17.634 40.072
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 31.4150 13.0584 2.406 0.02 *
T12 2.7010 0.6266 4.311 8.04e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 20.5 on 48 degrees of freedom
Multiple R-squared: 0.2791, Adjusted R-squared: 0.2641
F-statistic: 18.58 on 1 and 48 DF, p-value: 8.041e-05plot(O3~T12, data=ozone)
T12 <- seq(min(ozone[,"T12"]), max(ozone[,"T12"]), length = 100)
grille <- data.frame(T12)
ICdte <- predict(reg, new=grille, interval="conf", level=0.95)
matlines(grille$T12, cbind(ICdte), lty = c(1,2,2), col = 1)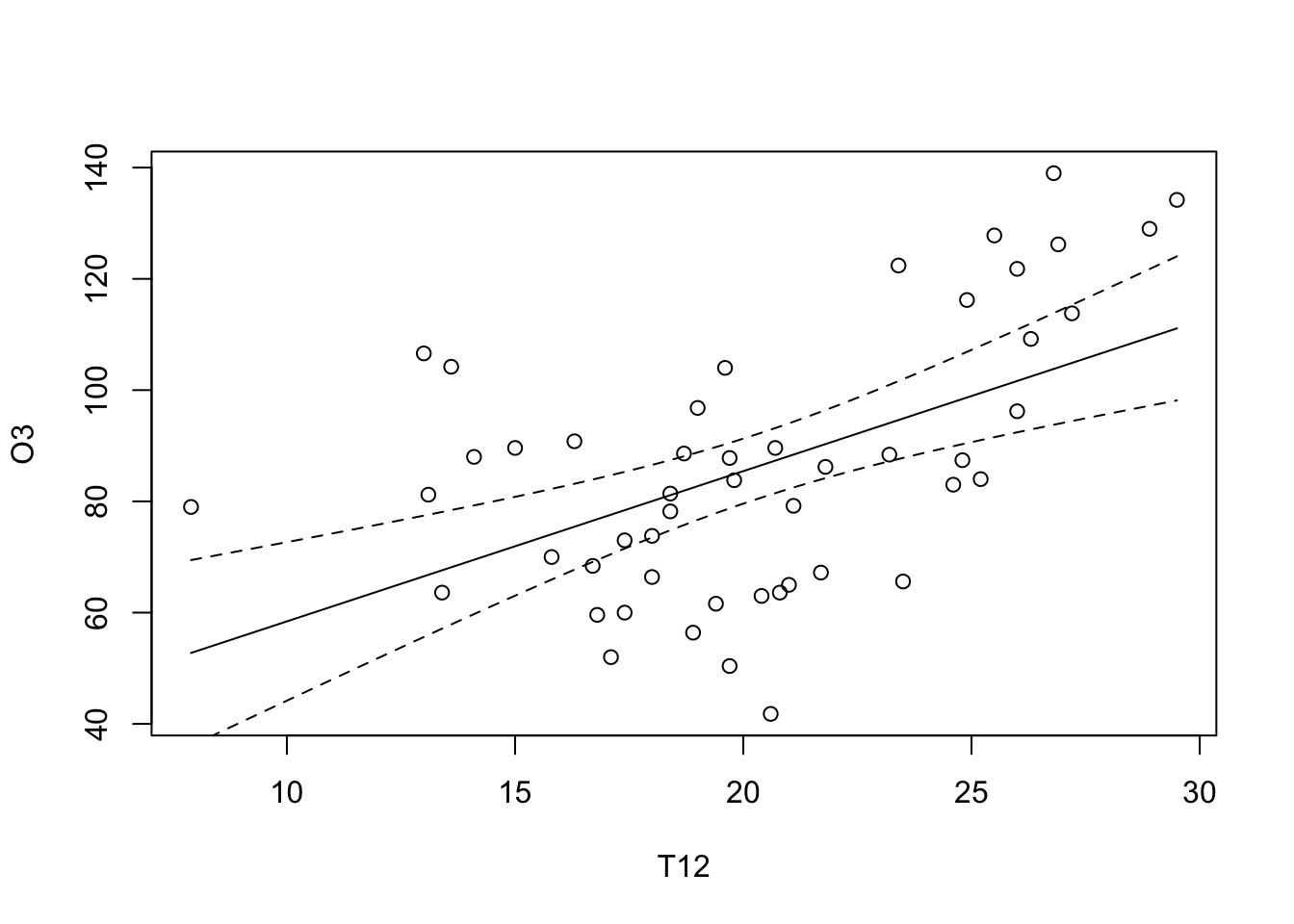
plot(O3~T12, data = ozone, ylim = c(0,150))
T12 <- seq(min(ozone[,"T12"]), max(ozone[,"T12"]), length = 100)
grille <- data.frame(T12)
ICdte <- predict(reg, new=grille, interval="conf", level=0.95)
ICprev <- predict(reg, new=grille, interval="pred", level=0.95)
matlines(T12, cbind(ICdte,ICprev[,-1]), lty=c(1,2,2,3,3), col=1)
legend("topleft", lty=2:3, c("Y","E(Y)"))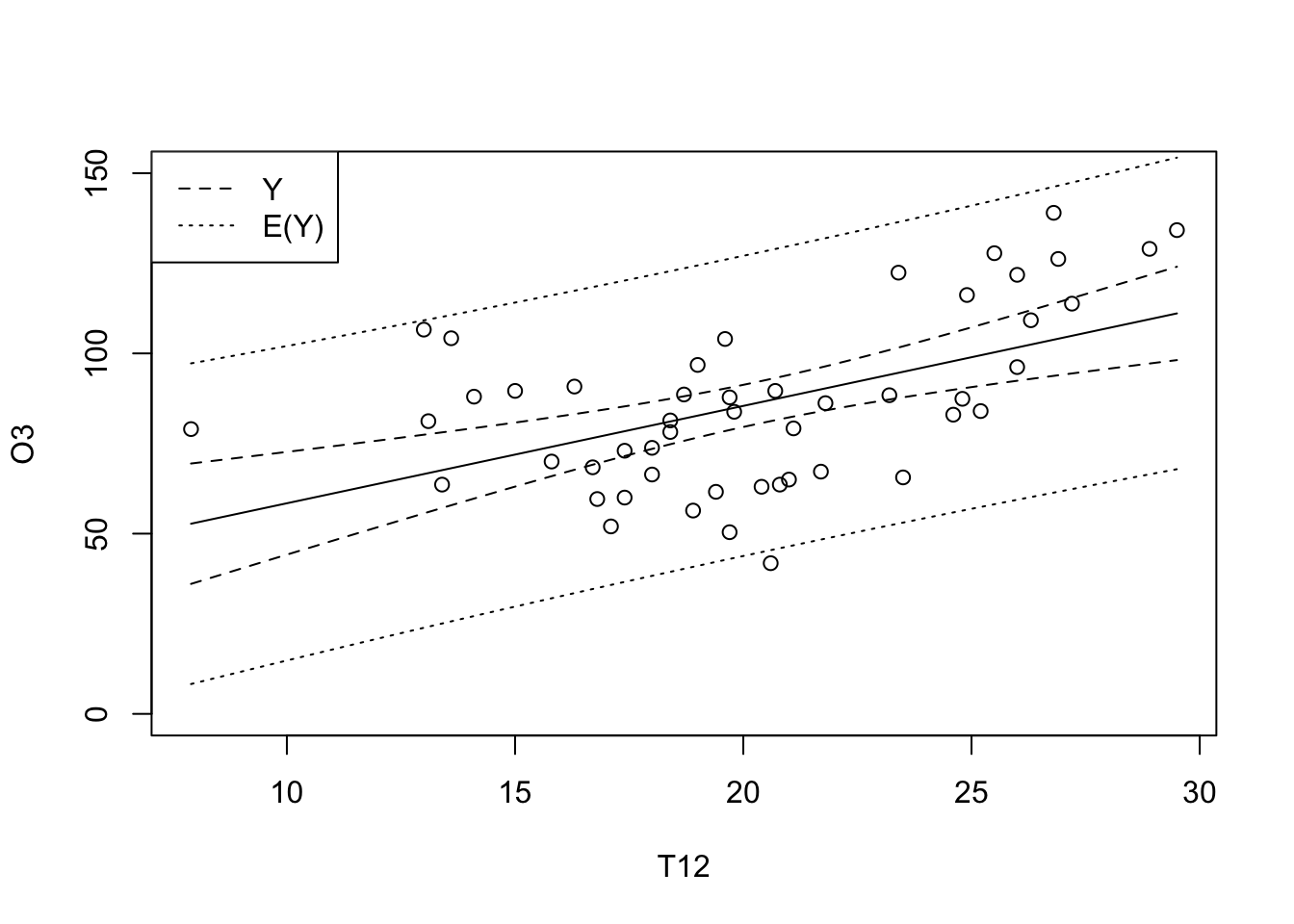
IC <- confint(reg, level = 0.95)
IC 2.5 % 97.5 %
(Intercept) 5.159232 57.67071
T12 1.441180 3.96089library(ellipse)
plot(ellipse(reg, level=0.95), type = "l", xlab = "", ylab = "")
points(coef(reg)[1], coef(reg)[2], pch = 3)
lines(IC[1,c(1,1,2,2,1)], IC[2,c(1,2,2,1,1)], lty = 2)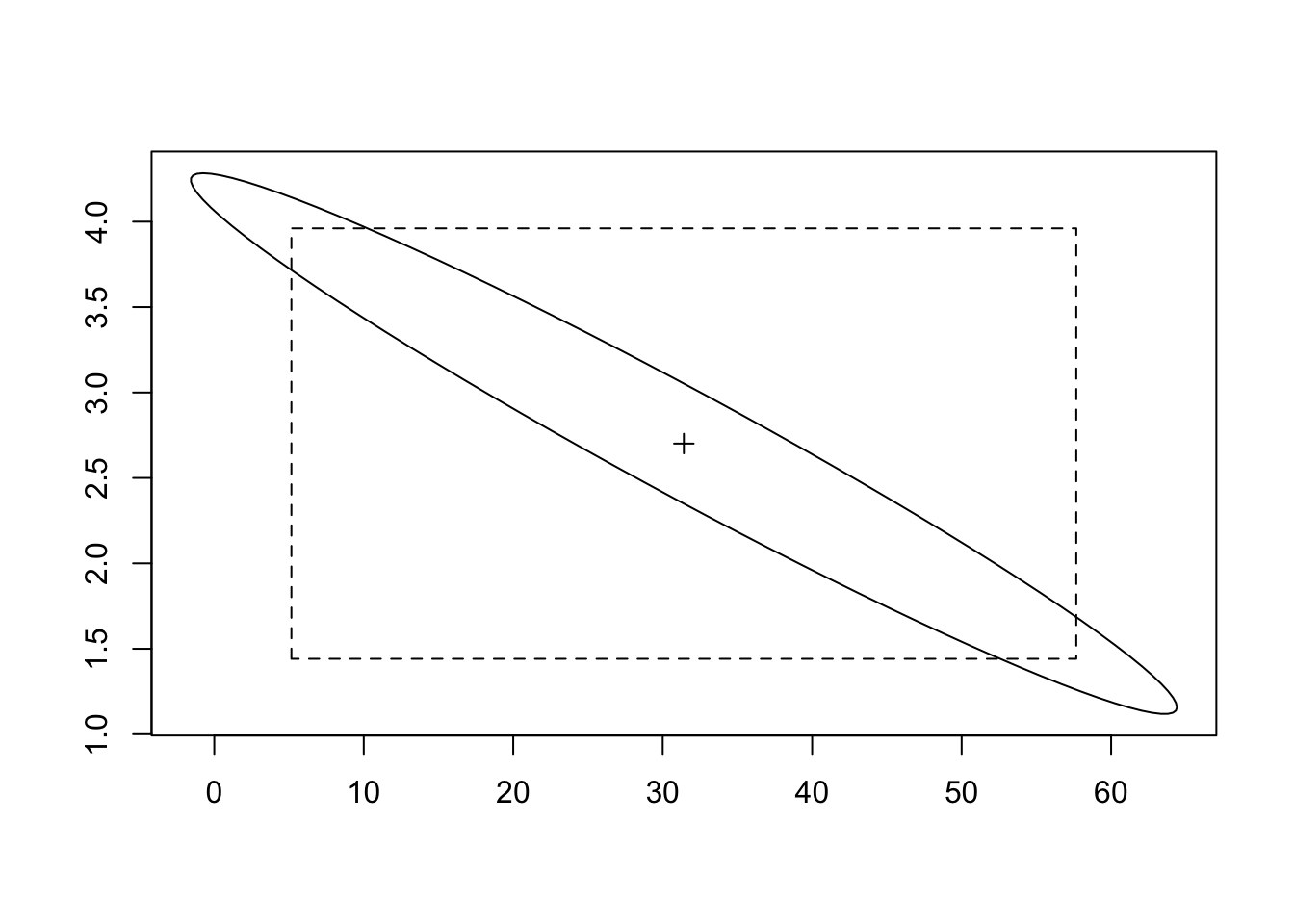
eucalypt <- read.table("../donnees/eucalyptus.txt", header = T, sep = ";")
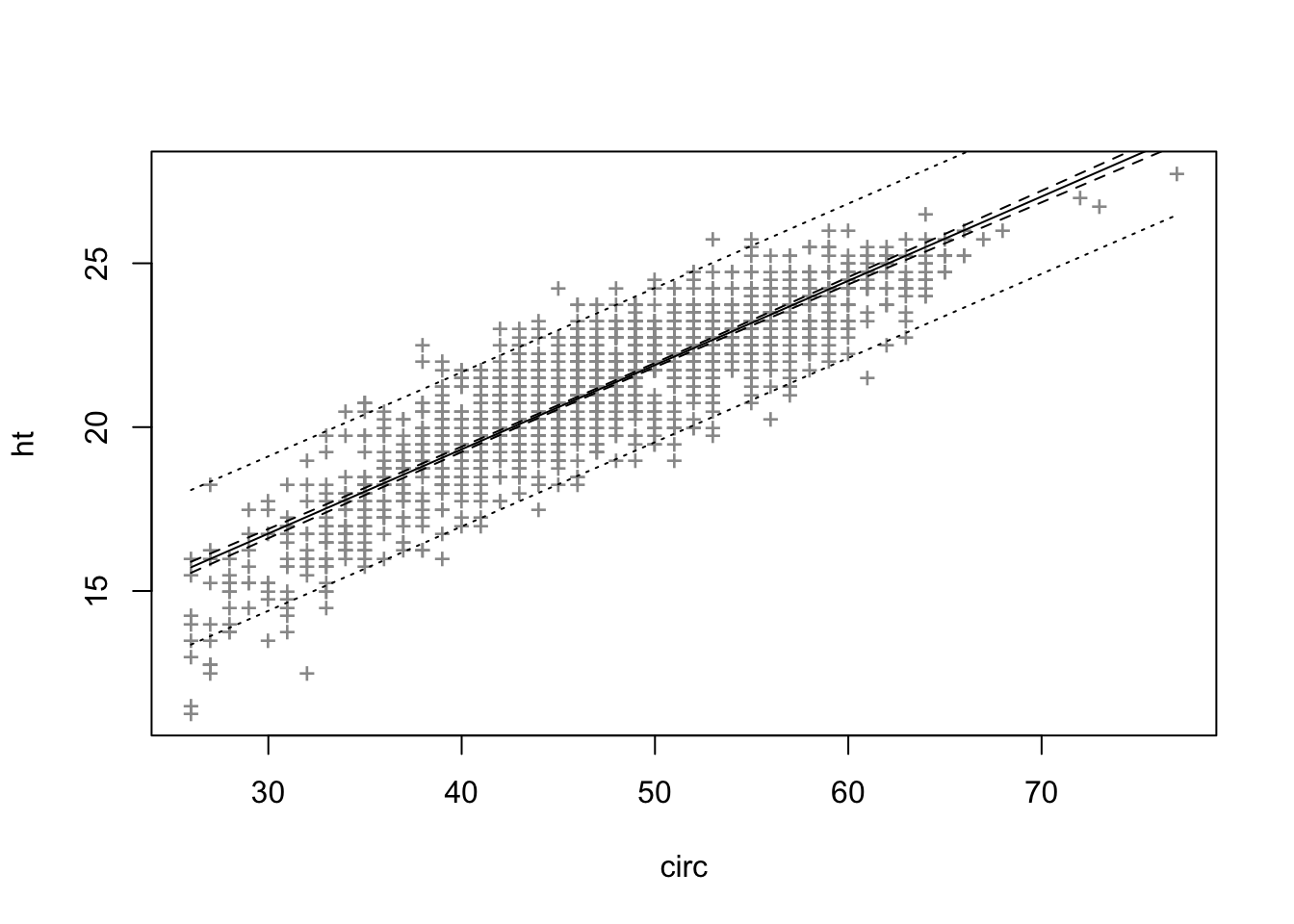
plot(ht~circ, data = eucalypt, xlab = "circ", ylab = "ht")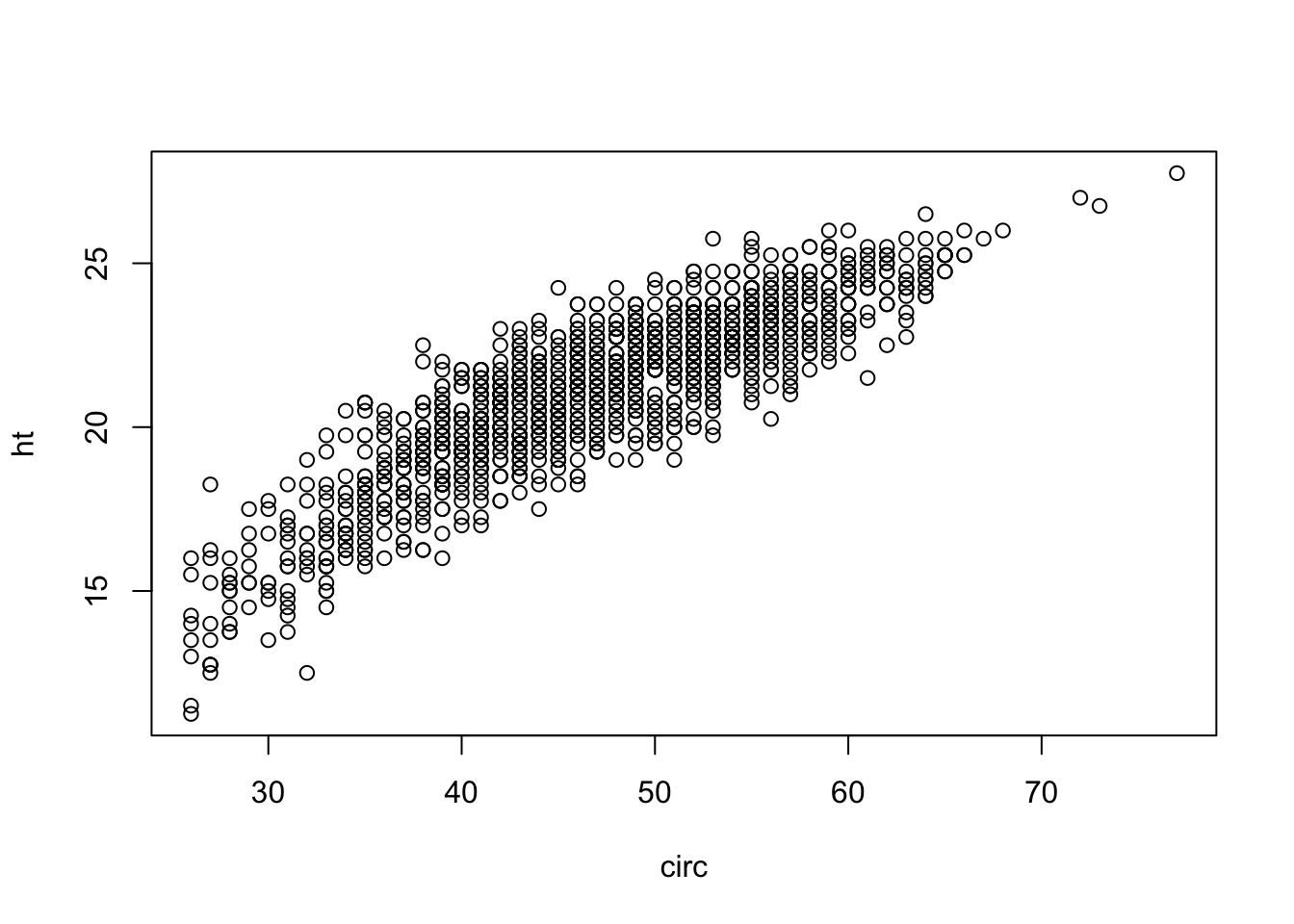
reg <- lm(ht~circ, data = eucalypt)
summary(reg)
Call:
lm(formula = ht ~ circ, data = eucalypt)
Residuals:
Min 1Q Median 3Q Max
-4.7659 -0.7802 0.0557 0.8271 3.6913
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 9.037476 0.179802 50.26 <2e-16 ***
circ 0.257138 0.003738 68.79 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.199 on 1427 degrees of freedom
Multiple R-squared: 0.7683, Adjusted R-squared: 0.7682
F-statistic: 4732 on 1 and 1427 DF, p-value: < 2.2e-16plot(ht~circ, data = eucalypt, pch = "+", col = "grey60")
grille <- data.frame(circ = seq(min(eucalypt[,"circ"]),
max(eucalypt[,"circ"]), length = 100))
ICdte <- predict(reg, new=grille, interval="confi", level=0.95)
matlines(grille$circ, ICdte, lty = c(1,2,2), col = 1)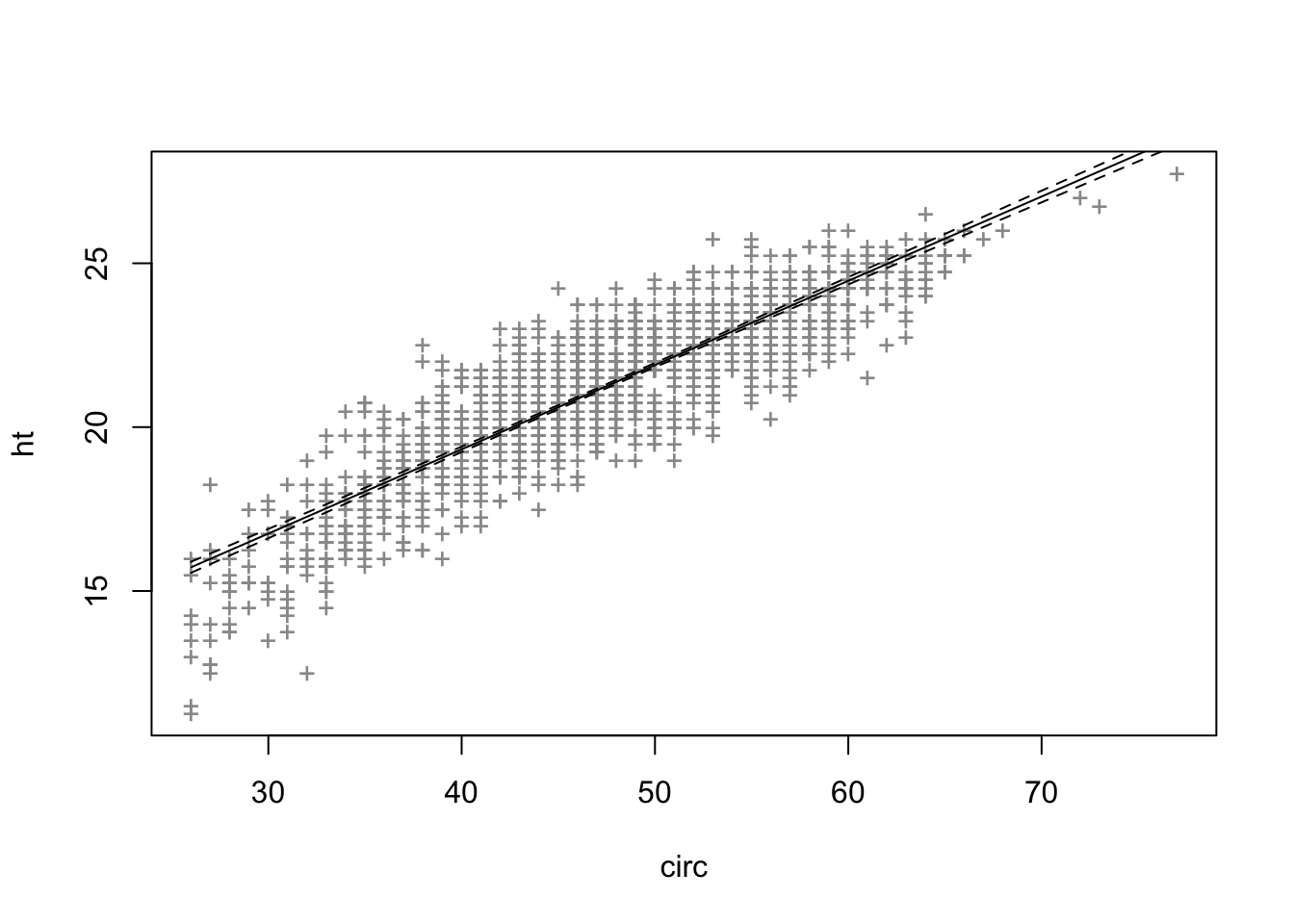
plot(ht~circ, data = eucalypt, pch = "+", col = "grey60")
circ <- seq(min(eucalypt[,"circ"]),max(eucalypt[,"circ"]), len = 100)
grille <- data.frame(circ)
Cdte <- predict(reg, new=grille, interval="conf", level=0.95)
ICprev <- predict(reg, new=grille, interval="pred", level=0.95)
matlines(circ, cbind(ICdte,ICprev[,-1]),lty=c(1,2,2,3,3), col=1)