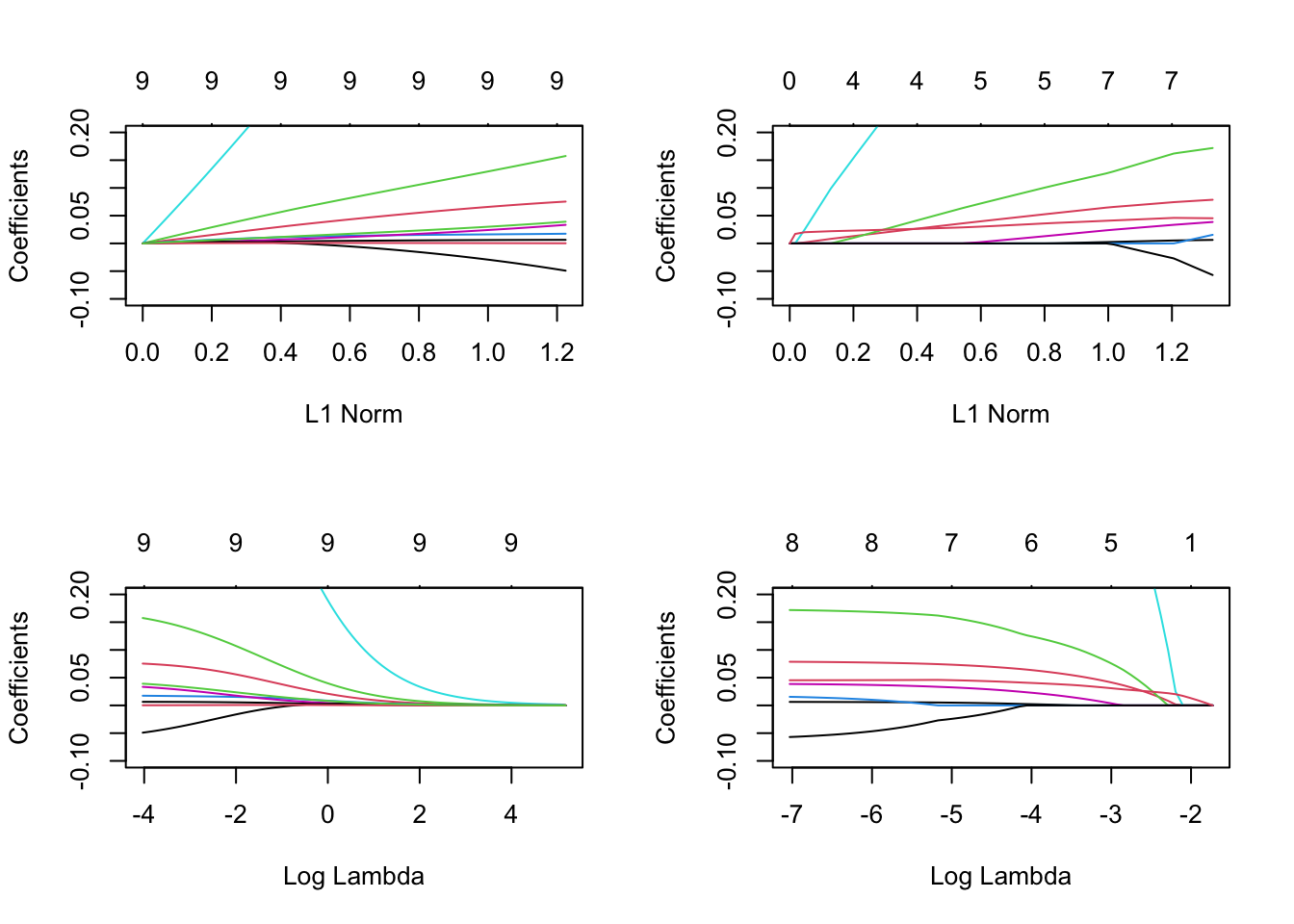
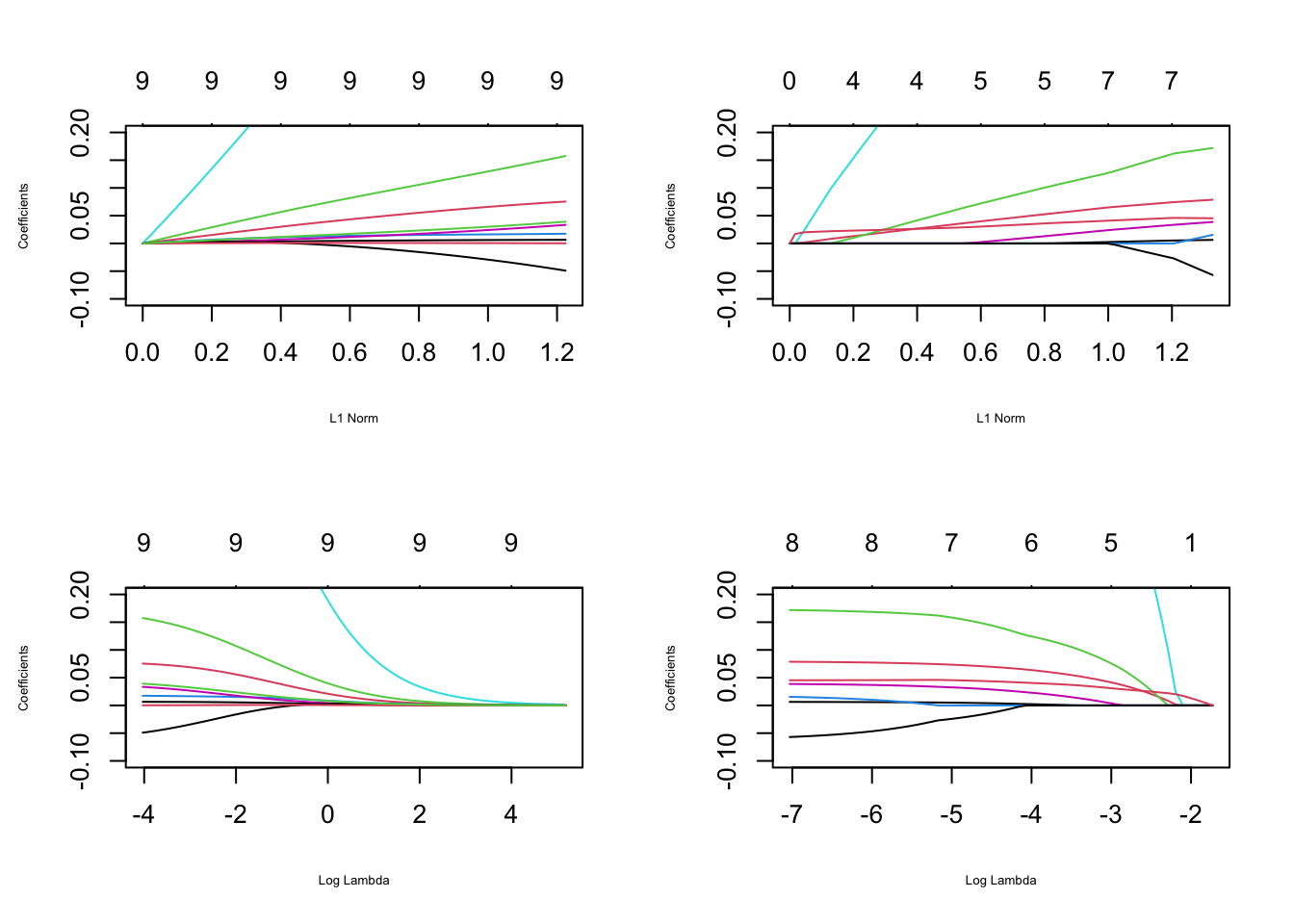
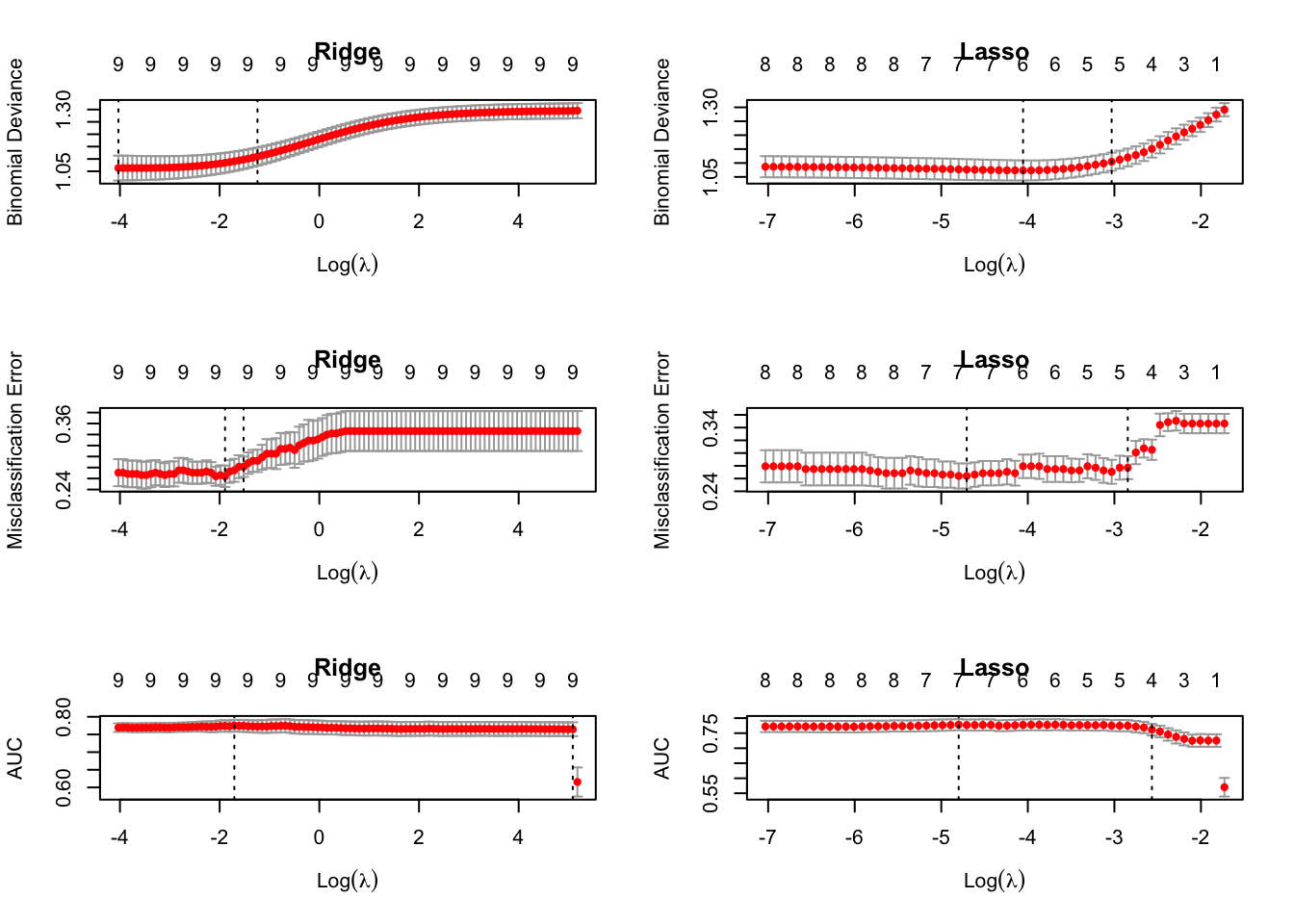
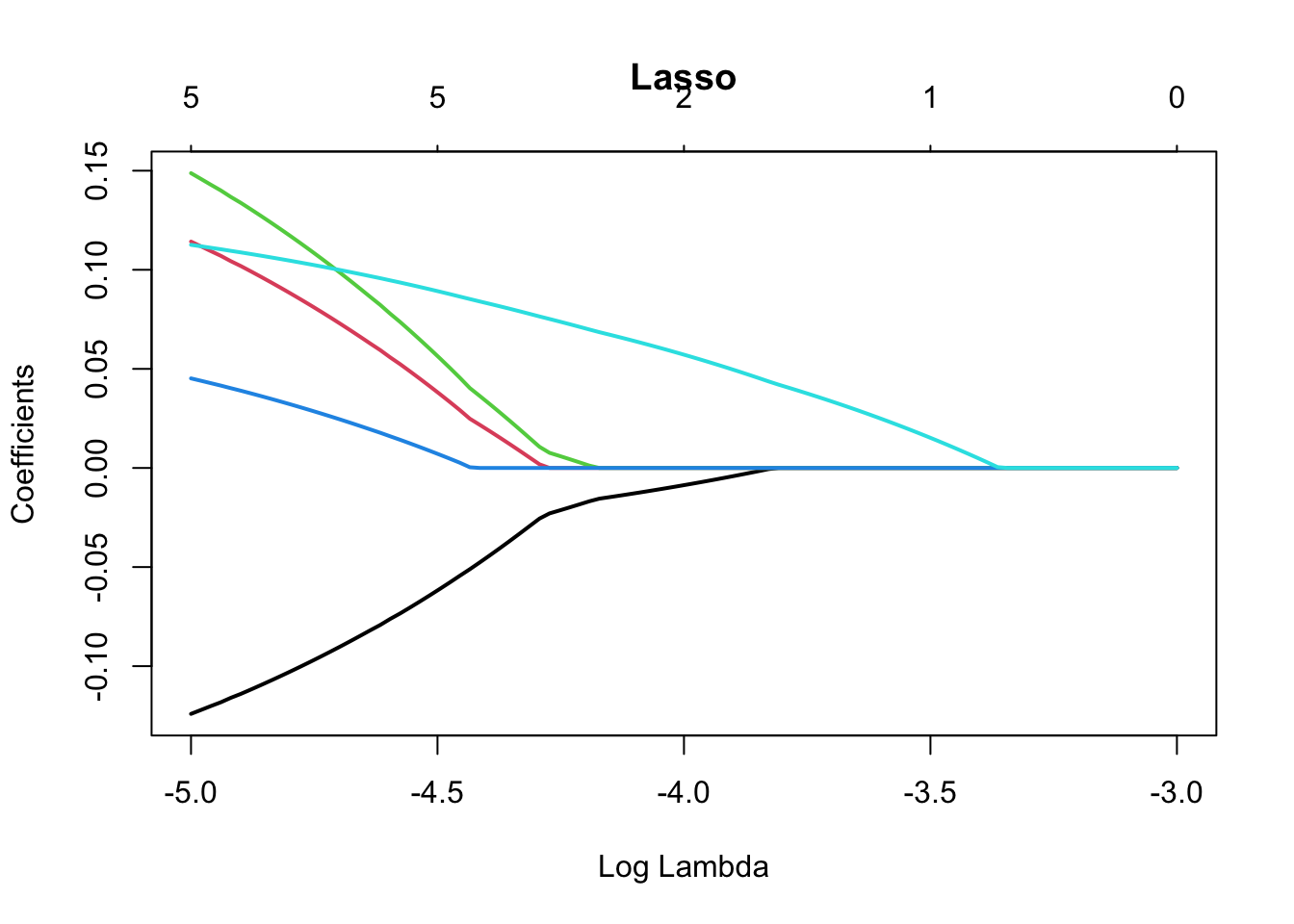
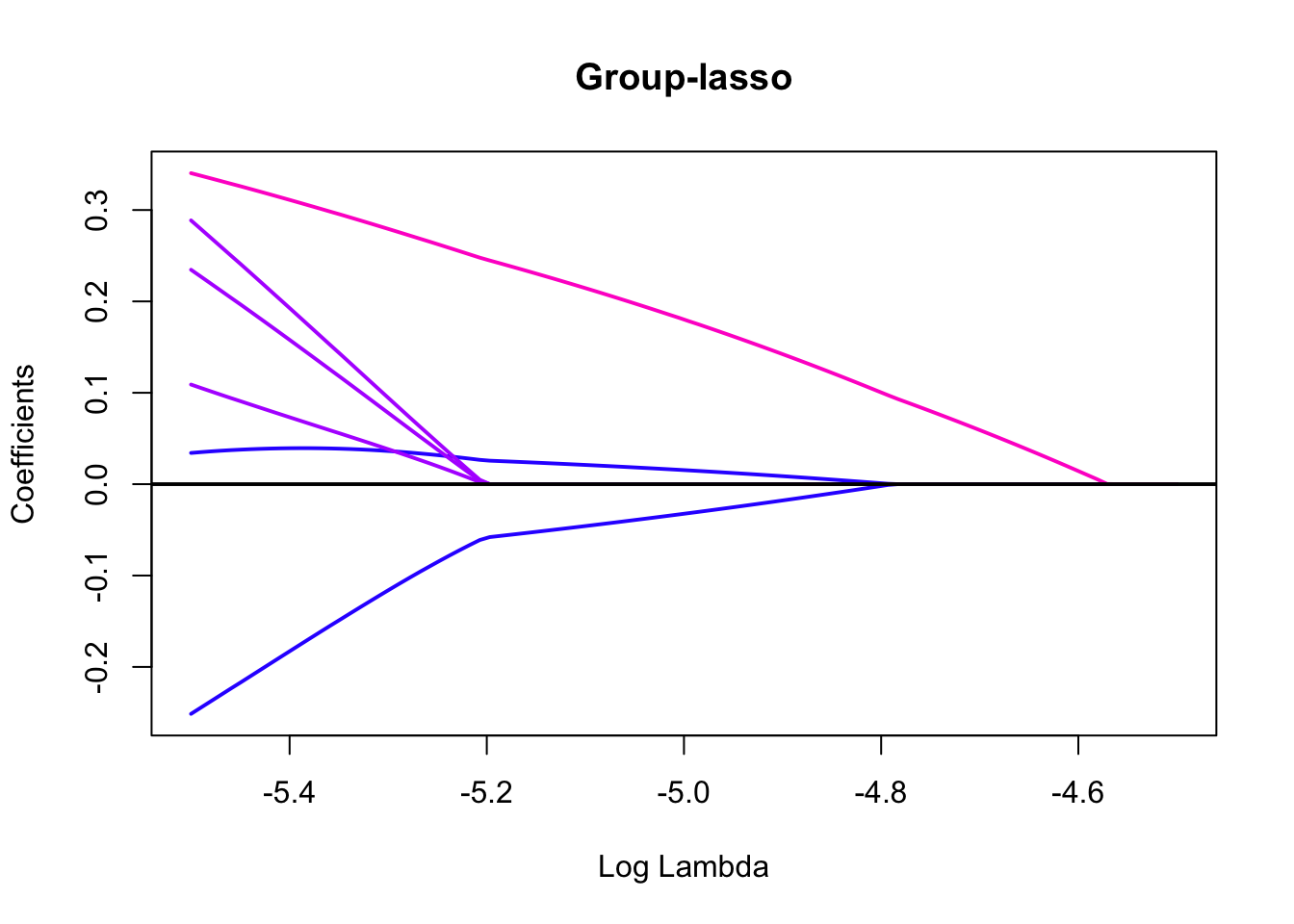
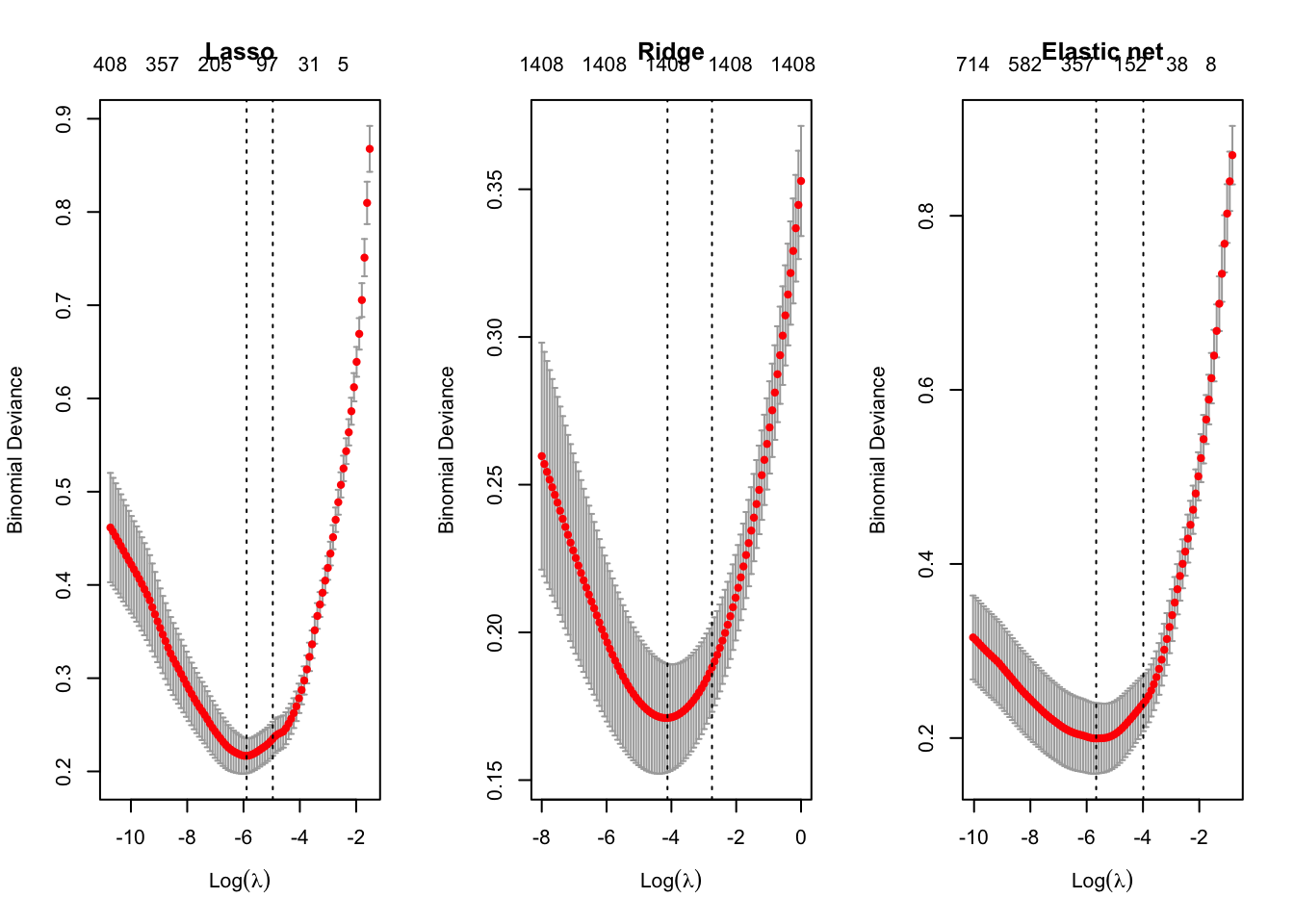
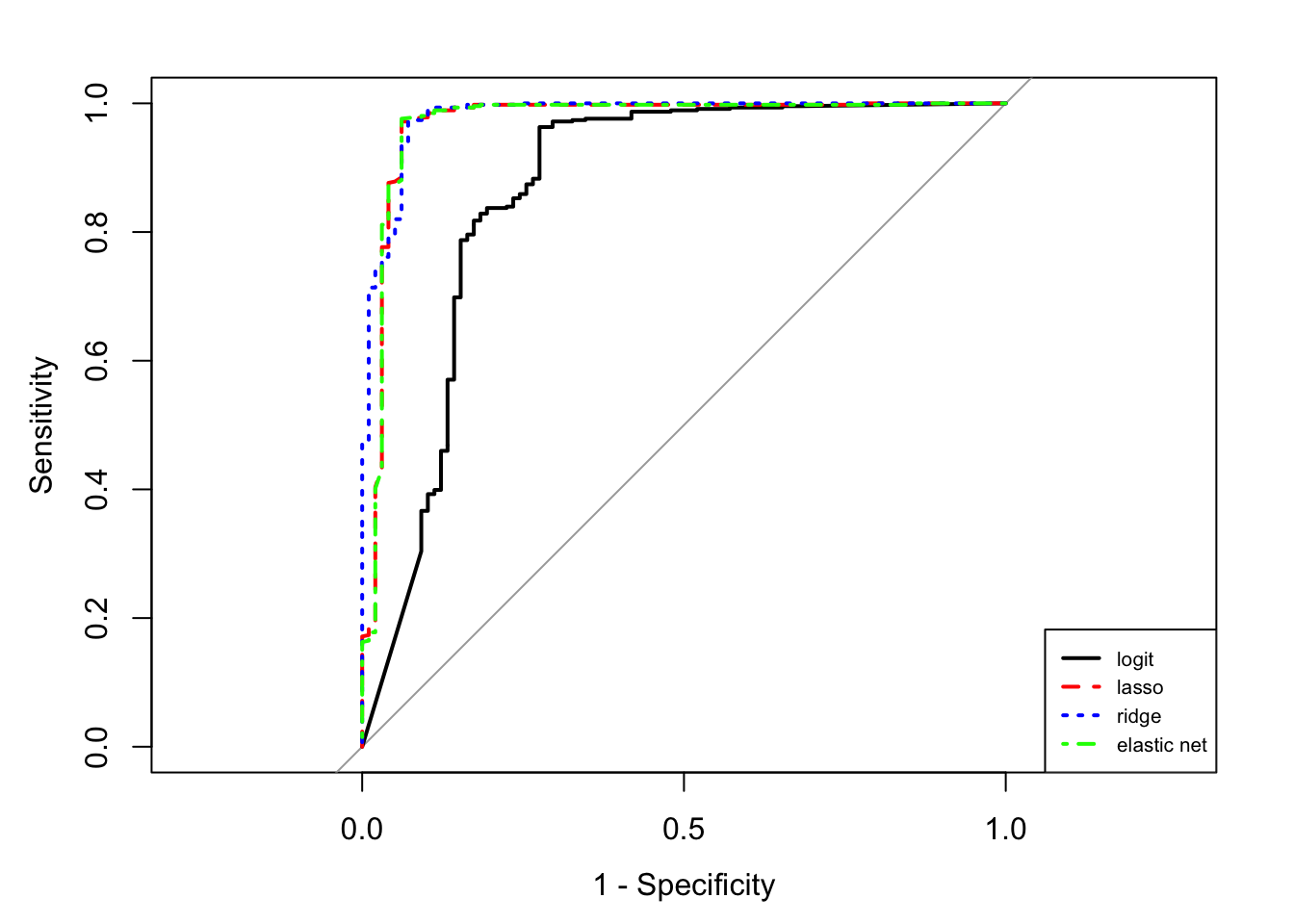
library(pROC)
roc.ad <- roc(obs~logit+lasso+ridge+en,data=score)
couleur <- c("black","red","blue","green")
mapply(plot,roc.ad,col=couleur,lty=1:4,add=c(F,T,T,T),lwd=2,legacy.axes=TRUE)
logit lasso ridge en
percent FALSE FALSE FALSE FALSE
sensitivities numeric,329 numeric,276 numeric,515 numeric,266
specificities numeric,329 numeric,276 numeric,515 numeric,266
thresholds numeric,329 numeric,276 numeric,515 numeric,266
direction "<" "<" "<" "<"
cases numeric,461 numeric,461 numeric,461 numeric,461
controls numeric,98 numeric,98 numeric,98 numeric,98
fun.sesp ? ? ? ?
auc 0.8635619 0.9695648 0.9805879 0.969587
call expression expression expression expression
original.predictor numeric,559 numeric,559 numeric,559 numeric,559
original.response factor,559 factor,559 factor,559 factor,559
predictor numeric,559 numeric,559 numeric,559 numeric,559
response factor,559 factor,559 factor,559 factor,559
levels character,2 character,2 character,2 character,2
predictor.name "logit" "lasso" "ridge" "en"
response.name "obs" "obs" "obs" "obs"
legend("bottomright",legend=c("logit","lasso","ridge","elastic net"),col=couleur,lty=1:4,lwd=2,cex=0.65)