ozone <- read.table("../donnees/ozone.txt", header = T, sep = ";")
modele3 <- lm(O3 ~ T12 + Vx + Ne12, data = ozone)
resume3 <- summary(modele3)
coef3 <- coef(resume3)
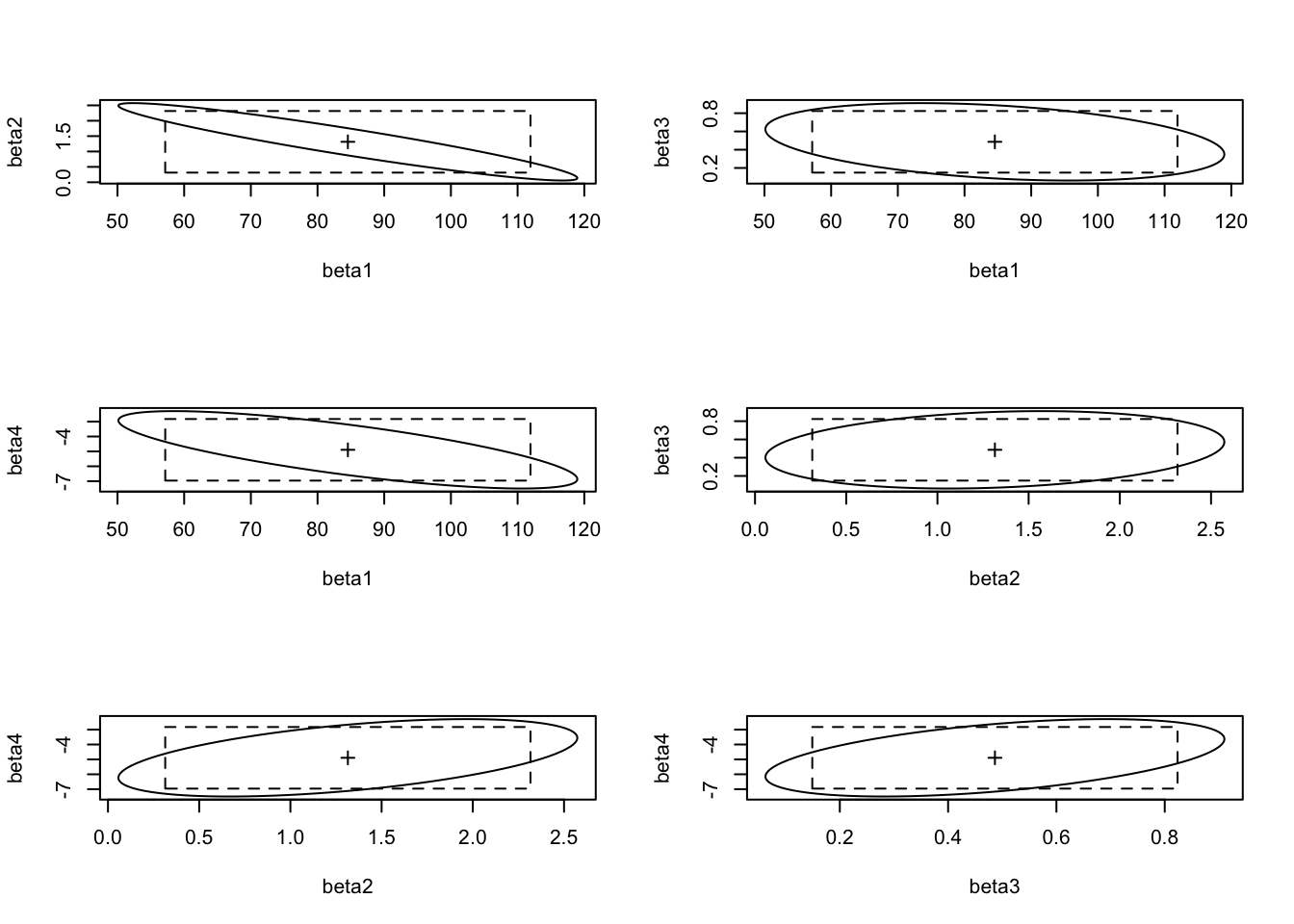
IC3 <- t(confint(modele3, level = 0.95))
IC3 (Intercept) T12 Vx Ne12
2.5 % 57.15842 0.3138112 0.1491857 -6.960609
97.5 % 111.93625 2.3162807 0.8237055 -2.826137